A research team is working on ways to produce 3D structures using DNA.
DNA is the foundation of life, as DNA molecules carry digital information that is used by cells to reproduce and develop all the functional biology required for life.
DNA can do this because it contains four molecules that can be easily bonded into complex structures. The molecules, adenine (A), cytosine (C), guanine (G), and thymine (T), can be arranged in “ladder” forms that are self-assembling.
The researchers wanted to leverage these properties to enable the production of nanostructures for a variety of applications. They call this science “DNA origami”, after the Japanese practice of folding paper into highly complex shapes. In turn, this is all about “folding” molecules into specific complex shapes.
Why do this? They explain the potential application types:
“Many three-dimensional (3D) DNA origami shapes can also be designed in such a way as to serve as a separating barrier between encapsulated internal moieties and an external environment. Emerging applications using encapsulating 3D DNA nanostructures have affected areas such as nanoparticle synthesis, low-volume reactors, templated assembly, molecular transport, or drug delivery. These applications often demand rigid, hollowed structures or, in other words, capsule-like functionality.”
The researchers devised a system they named “DNAxiS” to produce arbitrary geometries in DNA.
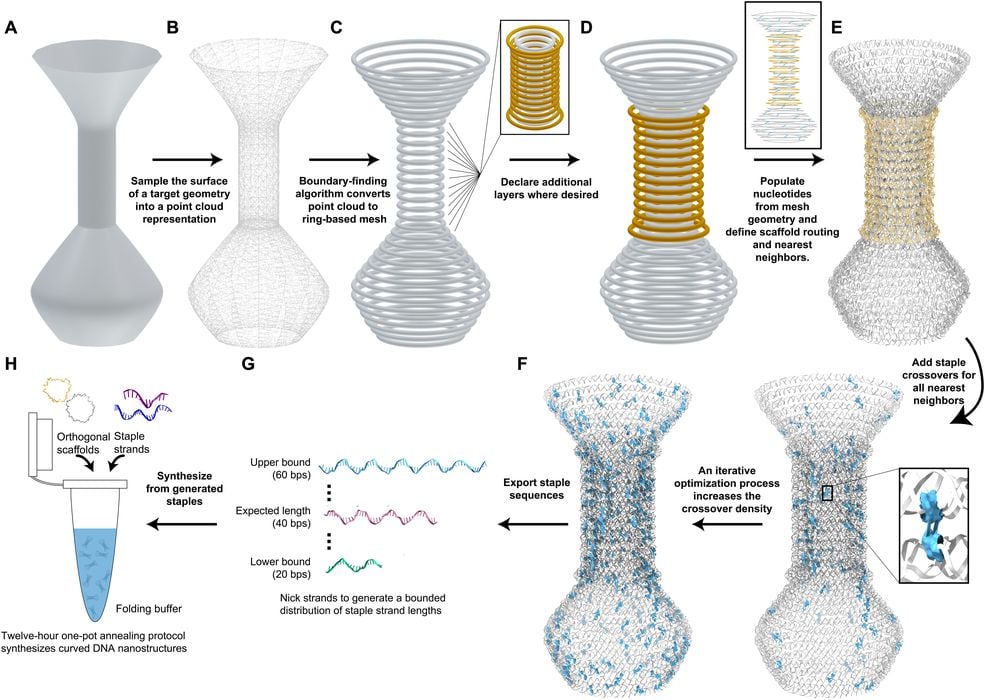
The process begins in the same way any 3D print process does: by designing a geometry in a CAD tool, and exporting it to a mesh format.
Their specialized software then decomposes the geometry into structures that can be made using the DNA bonding properties. Here is the process overview:
- User input is a 3D model in STL file format generated in the user’s graphics design software of choice
- The vertices of the STL build a point cloud that is upsampled to avoid gaps when extracting the shape’s outline
- A circle-based mesh is extracted from the point cloud
- The structure can be made selectively multilayer by adding rings outward from the starting mesh
- A helical twist is calculated from the circumference and used to convert each circle of the mesh into a DNA helix ring
- Crossovers are densely applied upon the template using either a greedy algorithm or a simulated annealing algorithm
- Conventional scaffold sequences are applied to generated corresponding staple sequences within specified length bounds
- Staple sequences are annealed with the corresponding scaffolds, sometimes multiple orthogonal sequences as needed, in a one-pot reaction to produce DNA nanostructures of the designed shape
The system was able to create quite a variety of basic shapes, and as readers will suspect, these could be linked together to create even more complex structures.
While this research intends on developing the technology further to be used in biomedical applications, one wonders whether similar approaches could be used to develop a kind of molecular 3D printing process for more general and larger-scale structures.
Via Science
